Extracting Drug-Drug Interactions (DDI) interactions from bio-medical literature.
 DDI example
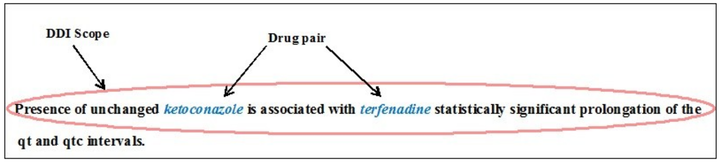
DDI exampleText mining techniques are useful in extracting hidden information about the biomedical interactions such as Protein-Protein, Drug-Drug and Protein-Drug. Recently, there is an increased interest in automated methods due to the vast growth in the volume of published text regarding biomedical interactions. This work mainly focuses on extraction of Drug-Drug interactions (DDIs) in biomedical research articles from well-known databases such as DrugBank and MedLine. Extracting such information manually from literature is very time and resource consuming. The goal of this research is to find automated techniques to extract interactions from unstructured biomedical text. The proposed approach is developed based on the features extracted using natural language processing (NLP) techniques such as bag-of-words approach, tokenization and so on. This allows a user to input a bio-medical text about drug interactions and through a series of steps which includes feature extraction techniques and use of classification algorithms, it will output automatically whether there is an interaction between the two drugs given.
This uncomplicated and easy to implement set of features are combined into a feature vector which is used to train a machine learning model. The effectiveness of the proposed approach was measured by conducting several experiments on the “DDI Extraction 2013” corpus. The system showed encouraging F-measure value of 76.9%.